Maura Barbisin,1 Ph.D.; Rixun Fang,1 Ph.D.; Cristin E. O’Shea,1 B.S.; Lisa M. Calandro,1 M.P.H.; Manohar R. Furtado,1 Ph.D.; and Jaiprakash G. Shewale,1 Ph.D.
Developmental Validation of the Quantifiler0002 Duo DNA Quantification Kit for Simultaneous Quantification of Total Human and Human Male DNA and Detection of PCR Inhibitors in Biological Samples*
ABSTRACT: The Quantifiler0003 Duo DNA Quantification kit enables simultaneous quantification of human DNA and human male DNA as well as detection of inhibitors of PCR in a single real-time PCR well. Pooled human male genomic DNA is used to generate standard curves for both human (ribonuclease P RNA component H1) and human male (sex determining region Y) specific targets. A shift in the cycle threshold (CT) values for the internal positive control monitors the presence of PCR inhibitors in a sample. The assay is human specific and exhibits a high dynamic range from 0.023 to 50 ng ⁄ lL. In addition, the multiplex assay can detect as little as 25 pg ⁄ lL of human male DNA in the presence of a 1000-fold excess of human female DNA. The multiplex assay provides assessment of the DNA extract and guidance for the selection of the appropriate AmpF‘STR0003 Amplification Kit to obtain interpretable short tandem repeat profiles.
KEYWORDS: forensic science, DNA quantification, real-time PCR, DNA analysis, ribonuclease P RNA component H1, sex determining region Y, human DNA, human male DNA, DNA typing
Forensic DNA analysis is targeted to obtain a short tandem repeat (STR) profile from an evidence sample, which is then compared with the STR profiles from reference samples collected from the victim and the suspect to determine the contribution to the evidence sample (1,2). STR genotyping systems such as Identifiler0003, Profiler Plus0003, COfiler0003, SGM Plus0003, MiniFilerTM, and Yfiler0003 kits are commercially available. The genotyping protocol, in general, involves extraction of DNA from the biological sample, quantification of the DNA, amplification for STR loci, and fragment analysis on a Genetic Analyzer. Quantification of human DNA in a forensic sample, which often contains nonhuman DNA, is an important step during STR profiling because the STR genotyping systems, unlike detection or single nucleotide polymorphism (SNP) assays, are sensitive to the quantity of DNA used in the PCR reaction: too little DNA may produce partial profiles whereas too much may produce off-scale data. For the forensic analyst, it is imperative to obtain an interpretable STR profile from forensic evidence samples, which often are in limited amount, and therefore, a reliable quantification method is vital. Hybridization-based quantification methods, e.g., Quantiblot0003, that are traditionally used for the quantification of DNA in forensic samples are generally considered time-consuming, labor-intensive, and not suitable for automation (3). Further, it is difficult to predict the amplitude of the STR profile because of the difference in the sensitivity of quantification methods and STR 1
Applied Biosystems, 850 Lincoln Centre Drive, Foster City, CA 94404. *Part of the work was presented at the 59th Annual Meeting of the American Academy of Forensic Sciences, in San Antonio, TX, February 19–24, 2007, and at the 18th International Symposium on Human Identification, Hollywood, CA, October 2007. Some of the data is also presented in the Quantifiler0003 Duo DNA Quantification Kit User Manual. Received 19 Jan. 2008; and in revised form 3 April 2008; accepted 20 April 2008.
0004 2009 Applied Biosystems Journal compilation 0004 2009 American Academy of Forensic Sciences
genotyping systems. Real-time PCR assays like the Quantifiler0003 Human DNA Quantification Kit and Quantifiler0003 Y Human Male DNA Quantification Kit have proved very useful (4). Real-time PCR assays for quantification of human DNA offer several advantages over the traditional hybridization assays such as: (i) specificity for a certain target in the genome because of the careful assay design; (ii) ability to detect few copies of target DNA; (iii) quantitative relationship between the amount of target template and the amount of PCR product accumulated at any given cycle prior to reaching saturation; (iv) greater dynamic range; (v) multiplexing capabilities; (vi) easy to adopt; and (vii) automatable for high throughput. A forensic evidence sample is often a mixture of human male and female DNA. Further, a forensic biological sample may be exposed to different environmental insults leading to DNA degradation and contamination with compounds that inhibit the PCR. Thus, it is desirable for the forensic analyst to have useful information about the forensic evidence sample prior to amplification for STRs. Real-time quantification assays can provide: (i) mixture ratio of human and human male DNA for choosing between autosomal and Y STR profiling based on the extent of the mixture ratio; (ii) presence of PCR inhibitors so that DNA extracts containing PCR inhibitors may be repurified prior to STR profiling; and (iii) quantification of human female and human male DNA useful for determining the quantity of extract to be used for amplification for different STR multiplex systems. To obtain the quantity of human and human male DNA in a sample using Quantifiler0003 Human and Y kits, it is necessary to run two separate quantification assays. This approach may consume a considerable amount of sample, which is often available in limited quantities, as well as time and reagents. To overcome these hurdles,
305
306
JOURNAL OF FORENSIC SCIENCES
multiplex real-time quantification assays have been described in recent years (5–8). The assays described by Walker et al. (5), Horsman et al. (6), and Nicklas and Buel (7) enable an estimation of the mixture ratio of human male and female DNA but were not designed to detect the inhibitors of PCR in the sample. The assay described by Swango et al. (8) enables forensic analysts to obtain the mixture ratio as well as the detection of the inhibitors of PCR. However, the human nuclear DNA amplification target THO1 (human tyrosine hydroxylase gene on chromosome 11) spans the polymorphic STR region (8). THO1 is a commonly used locus for human identification in forensic laboratories (2). First, the incorporation of such polymorphic STR target in the quantification assay is discouraged to avoid possible contamination incidences in the laboratory. Second, the length of the amplicon would vary by 44 nucleotides since alleles ranging from 3 to 14 repeat units have been characterized in different human population groups (for references see Short Tandem Repeat DNA Internet DataBase compiled by NIST available at http://www.cstl.nist.gov/biotech/strbase/str_ TH01.htm). Though the detection probe is designed outside the polymorphic region, the possibility of variation in the efficiency of the assay due to the variation in the length of the amplicon can not be ruled out. We describe a multiplex TaqMan0003 real-time PCR assay, the Quantifiler0003 Duo DNA Quantification Kit, for simultaneous quantification of human nuclear and human male DNA as well as detection of the presence of PCR inhibitors in a biological sample. The developed assay enables the assessment of the biological samples for downstream STR profiling. Materials and Methods Pooled human male genomic DNA used for generation of standard curves was obtained from EMD Biosciences Inc. (San Diego, CA). Genomic DNA from unknown individuals was obtained from Biochain (Hayward, CA), Sigma Chemical Company (St. Louis, MO), Promega (Madison, WI), or Serological Research Institute (Richmond, CA). Nonhuman samples were obtained as purified DNA from BIOS Laboratories, Inc. (New Haven, CT), Pel-Freez Biologicals (Rogers, AR), and American Type Culture Collection (Manassas, VA). Oligonucleotides, TaqMan0003 probes, Quantifiler0003 human DNA quantification kit, Quantifiler0003 Y male DNA quantification kit, AmpF‘STR kits, 7500 Real-time PCR System, 3130 Genetic Analyzers and associated software were from Applied Biosystems (Foster City, CA). All other chemicals used in this study were of analytical grade. Extraction and Quantitation of DNA The DNA from anonymous donor samples (blood, saliva and semen, either liquid or stains, and buccal swabs) was extracted by using standard phenol–chloroform (9), or BloodPrep0003 DNA Chemistry and the ABI PRISM0003 6100 Nucleic Acid PrepStation (Applied Biosystems) procedures. The quantity of DNA was determined by Quantifiler0003 Duo, Quantifiler0003 Human, and Quantifiler0003 Y Human Male DNA Quantification Kits (Applied Biosystems). Real-Time PCR Amplification Real-time PCR amplification reactions contained 10.5 lL of Primer-Probe Mix, 12.5 lL of Master Mix, and 2.0 lL of DNA sample. The Primer-Probe Mix contained forward and reverse primers and TaqMan0003 probes for ribonuclease P RNA component H1 (RPPH1), sex determining region Y (SRY), and internal PCR
control (IPC) targets. The IPC template, a synthetic polynucleotide, was cloned into a plasmid. The Master Mix contained Reference dye, dNTPs, dUTP, MgCl2, AmpliTaq0003 Gold DNA polymerase and preservatives in Tris-HCl, pH 8.0. Pooled human male genomic DNA at eight different concentrations (50, 16.7, 5.56, 1.85, 0.62, 0.21, 0.068, and 0.023 ng ⁄ lL) was amplified on each quantification run plate for generation of standard curves for RPPH1 and SRY targets. Amplification reactions were performed in a 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer’s instruction with conditions as follows: 500005C, 2 min; 950005C, 10 min; 40 cycles of 950005C, 15 sec and 600005C, 1.0 min. The data were analyzed using 7500 System sds Software v1.2.3 (Applied Biosystems) with a threshold value of 0.2. STR Analysis The samples were amplified with Identifiler0003, Yfiler0003, and MiniFilerTM kits using the procedure described in the User’s Manual for the respective kit. The amplified products were analyzed on a 3130xl Genetic Analyzer (Applied Biosystems) with GeneMapper0003 ID Software v3.2.1 (Applied Biosystems). Sensitivity Study Two human male genomic DNA samples, one pooled and the other single source, obtained from commercial sources were diluted to obtain concentrations of 20.0, 5.0, 1.0, 0.1, 0.05, 0.04, 0.03, 0.023, 0.0115, 0.00575, 0.002875, and 0.00144 ng ⁄ lL in 10 mM Tris buffer, pH 8.0 containing 0.1 mM ethylene diamine tetraacetic acid (EDTA). Each dilution was quantified in triplicate using the Quantifiler0003 Duo DNA Quantification Kit. Species Specificity The DNA from nonhuman biological species was either obtained commercially or purified in the laboratory. For some of these DNA samples, the sex of the donor animal was unknown. For some species, multiple donor animals were tested. Most of the reactions utilized 5.0 ng of input DNA. For a few reactions, 10 ng of input DNA was used. Precision and Accuracy One set of eight serial dilutions was prepared containing 50, 16.7, 5.56, 1.85, 0.62, 0.21, 0.068, and 0.023 ng ⁄ lL of the human male DNA standard present in the Quantifiler0003 Duo DNA Quantification Kit. Six reaction plates were set up and each of them contained 10 replicates of the eight dilutions. Two plates per instrument were run on three different 7500 Real-time PCR System instruments. The two runs were performed on two different days, using the same three 7500 Real-time PCR System instruments. For each dilution, the CT values for RPPH1, SRY, and IPC signals were recorded for all 60 reactions. Reproducibility Four male and one female genomic DNA samples were diluted from initial estimated concentrations to 20.0, 10.0, 1.0, 0.1, and 0.05 ng ⁄ lL. All dilutions were made in 10 mM Tris buffer, pH 8.0 containing 0.1 mM EDTA. All samples and dilutions were run in triplicate using the Quantifiler0003 Duo Kit. Three runs were performed on different days. For each sample reaction, the CT values were obtained and the DNA quantities calculated.
BARBISIN ET AL. • QUANTIFILER0002 DUO KIT VALIDATION
307
Mixture Study
Degraded DNA
Mixture samples containing 0.2 ng ⁄ lL of human male DNA and varying amounts of female DNA were prepared. The ratio of male to female DNA in these samples was 1:0, 1:1, 1:5, 1:10, 1:20, and 0:1. The mixture samples were processed in triplicate using the Quantifiler0003 Duo DNA Quantification Kit to determine the concentration of total human DNA (RPPH1 target) and male DNA (SRY target). Using the quantification results from the RPPH1 human target, c. 1.0 ng of human genomic DNA from each sample was added to an Identifiler0003 kit reaction. Similarly, using the results from the SRY male target, c. 1.0 ng of human genomic DNA from each sample was added to a Yfiler0003 kit reaction. Another set of mixture samples containing 25 pg ⁄ lL of male DNA and increasing quantities of female DNA was prepared to obtain male to female DNA ratios of 1:0, 1:50, 1:100, 1: 200, 1:500, 1:800, 1:1000, and 0:1. The samples were processed in triplicate using the Quantifiler0003 Duo Kit to determine the concentration of total human genomic DNA (RPPH1 target) and male DNA (SRY target). In addition, based on the results from the SRY male target, c. 1.0 ng of human genomic DNA from each sample was added to the Yfiler0003 reaction.
One microgram of DNA (100 lL reaction at 10 ng ⁄ lL concentration) was treated for 20 min using varying quantities of the DNase I enzyme; 0.002, 0.01, 0.02, 0.05, 0.1, and 0.2 units. Samples were run on a 4% agarose gel for 25 min and visualized by staining with ethidium bromide to monitor the extent of degradation. The degraded DNA samples were processed with the Quantifiler0003 Duo kit to determine the quantity of amplifiable DNA at each level of degradation. Results obtained using the RPPH1 human target of the Quantifiler0003 Duo kit were used to calculate DNA input for STR analysis using Identifiler0003 and MiniFilerTM kits.
Calculation of Male to Female DNA Ratio The Quantifiler0003 Duo kit provides the quantity of human and human male DNA in biological samples. From these values, one can calculate the ratio of male and female DNA using the following equation: Male DNA:Female DNA Ratio Male DNA ðHuman DNA 0002 Male DNA) : ¼ Male DNA Male DNA or Male DNA:Female DNA Ratio ¼ 1 : ðHuman DNA 0002 Male DNAÞ=Male DNA All quantities in the above equations are ng ⁄ lL. This ratio determines the extent of the mixture, which is useful for making the choice of STR analysis method: autosomal STRs or Y-STRs. Inhibited Samples Human male genomic DNA was mixed with hematin to obtain final concentrations of 0, 2.5, 5.0, 7.5, 10, 12.5, 15, 17.5, 20, and 40 lM in the 25-lL quantification PCR. A second set of inhibited samples was prepared by addition of humic acid to obtain final concentrations of 0, 1.0, 2.0, 3.0, 3.75, 7.5, 11.25, 15, and 30 ng ⁄ lL in the 25-lL quantification PCR. The concentrations described here were final concentrations of respective inhibitor in 25-lL PCR when 2 lL of sample is added. The concentration of the inhibitor in the sample was, therefore, 12.5 times higher. Since the final concentration in the PCR was the contributing factor for the inhibition, the samples were named accordingly and the same nomenclature was used for both the quantification and the STR reactions for simplicity. Two microliters of each sample, containing c. 1.0 ng of DNA, was quantified in triplicate using the Quantifiler0003 Duo DNA Quantification Kit. Results obtained using the RPPH1 human target of the Quantifiler0003 Duo kit were used to calculate DNA input for STR analysis using Identifiler0003 and MiniFilerTM kits.
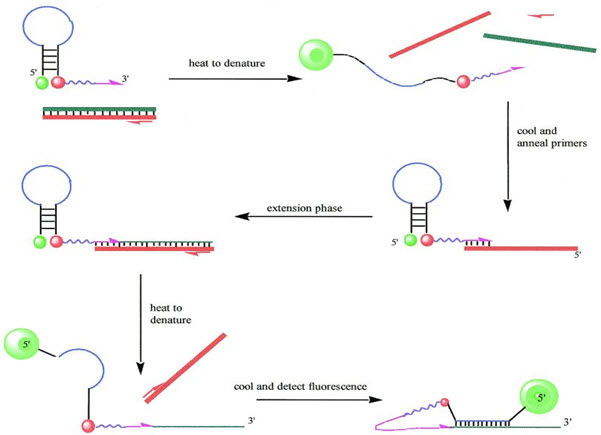
Case-Type Samples Saliva and blood samples were collected from four human male donors (donors a and b for saliva, donors c and d for blood). Semen was from one human male donor (donor e). Forensic-type samples were prepared by loading 50 lL of saliva on cotton swab, 5 lL of blood on fabric, 5 lL of blood on denim, 5 lL of blood on filter paper, 5 lL of blood spiked with inhibitors on fabric and 1 lL of semen on fabric (samples 1–8). The DNA was isolated by a phenol:chloroform extraction method. Extracted DNA was quantified in triplicate using the Quantifiler0003 Duo Kit. Results and Discussion Availability of STR profiling kits with targeted capabilities such as the Yfiler0003 kit for Y-STRs (10) and the MiniFilerTM kit for compromised samples (11) made it essential to assess the quality of a DNA extract in addition to the DNA quantification. The developed triplex assay comprises coamplification of the ribonuclease P RNA component H1 or RPPH1 gene (Gene ID 85495) for quantification of total human DNA, the sex determining region Y or SRY gene (Gene ID 6736) for quantification of human male DNA, and a synthetic nucleotide template sequence as an IPC in a single reaction. The human target RPPH1 is also known as H1 RNA or H1RNA and is located on chromosome 14 (location 14q11.2). The male target SRY is also referred as TDF or TDY and is located on chromosome Y (location Yp11.3). The genes are present at one copy per chromosome. Thus, two copies of RPPH1 and one copy of SRY are amplified during the quantification assay. The TaqMan0003 probes for the measurement of the human male, human, and IPC targets were labeled with FAMTM, VIC0003, and NEDTM dyes, respectively. The PCR mixture comprises two oligonucleotide primers and a TaqMan0003 probe specific for each target. The principle and mechanism of the real-time PCR quantification assay employing TaqMan0003 probes is described earlier (4). Briefly, the TaqMan0003 probes are labeled with a fluorescent reporter dye at the 5¢ end and a nonfluorescent quencher along with a minor groove binder (MGB) moiety at the 3¢ end. The extent of amplification for each target is determined by measuring the respective fluorescent dye released from the probe by 5¢ nuclease activity of the DNA polymerase during the extension phase of the PCR (12). A threshold for the fluorescence is set at the beginning of the exponential phase based on the initial cycles when little change in the fluorescence occurs. Cycle threshold (CT) value is the cycle at which the fluorescence signal crosses the threshold value. Thus, the lower the CT value, the higher the quantity of DNA. Real-time PCR assays can quantify the DNA present in a given well by measuring the CT value and comparing it with the standard curve CT values. The multiplex was optimized in silico to avoid interactions between the oligonucleotides and minimize the formation of primer-dimers. This was confirmed by laboratory testing. The primer and probe concentrations
308
JOURNAL OF FORENSIC SCIENCES
were optimized to ensure that the human male DNA was detected and quantified accurately in the presence of a large quantity of female DNA. The primers were selected and designed based on the published sequences to obtain 140, 130, and 130-bp fragments for the RPPH1, SRY, and IPC targets, respectively. The size of the amplicons in the Quantifiler0003 Duo kit is greater than the length of the hTERT (62 bp) and SRY (64 bp) targets in the Quantifiler0003 Human and Quantifiler0003 Y Human Male Quantification Kits, respectively (4). The size of the amplicons was increased in the Quantifiler0003 Duo kit to avoid incidences of over estimation of the quantity of DNA and to obtain a better correlation between the DNA quantification values and predictability of the STR profiles as described in this paper. Although the Quantifiler0003 Duo DNA Quantification Kit is not a DNA genotyping assay, it is intended for use before performing genotyping assays using AmpF‘STR0003 PCR Amplification kits. The developmental validation studies were performed following the revised validation guidelines provided by the Scientific Working Group on DNA Analysis Methods (SWGDAM) Guidelines (13). By testing the procedure with samples commonly encountered in forensic and parentage laboratories, the validation process clarifies attributes and limitations that are critical for sound data interpretation in casework. Standard Curves Linearity of quantification of the triplex assay was determined from the standard curves for human and human male targets generated by amplification of pooled human male genomic DNA at concentrations ranging from 0.023 to 50.0 ng ⁄ lL (Fig. 1). As expected, the CT value increased progressively with a decrease in the amount of human DNA. The CT values for the IPC increased by about 0.5–1.0 at higher concentrations of human DNA because of a slight PCR competition. A linear relationship between the CT values and the quantity of DNA template investigated was observed for both human and human male DNA. In general, the CT values for the SRY target were higher than those for the RPPH1 target. This observation is in concordance with the haploid nature of the SRY target and the diploid nature of the RPPH1 target in the human male sample. The CT values for the human target are higher than those for the male target in the multiplex assay reported by Horsman et al. (6); this could be attributed to lower amplification efficiency of the human target.
FIG. 1—Example of a typical standard curve: cycle threshold (CT) values for ribonuclease P RNA component H1 (RPPH1), sex determining region Y (SRY), and internal positive control (IPC) targets across the eight standard DNA concentrations (50, 16.7, 5.56, 1.85, 0.62, 0.21, 0.068, and 0.023 ng ⁄ lL).
TABLE 1—Sensitivity study: measured and expected quantities for two samples. Measured Quantity (ng ⁄ lL) Sample 1
Sample 2
Expected Quantity (ng ⁄ lL)
RPPH1
SRY
RPPH1
SRY
20 5 1 0.1 0.05 0.04 0.03 0.023 0.01150 0.00575 0.00288 0.00144
18.500 4.000 0.832 0.099 0.050 0.039 0.026 0.020 0.014 0.010 – –
19.540 4.330 0.909 0.111 0.048 0.053 0.033 0.022 0.009 0.007 0.000 0.006
20.910 4.943 0.802 0.096 0.056 0.038 0.038 0.022 0.015 0.010 0.006 –
20.383 4.800 0.751 0.108 0.058 0.039 0.031 0.033 0.016 – 0.007 –
SRY, sex determining region Y; RPPH1, ribonuclease P RNA component H1.
About 45 standard curves were generated on multiple instruments. The slope values for both standard curves ranged between )3.0 and )3.6 indicating amplification efficiency of 100 € 10% for both the targets. Similarly, the R2 values remained >98.0% allowing accurate quantification of each target simultaneously. Sensitivity Study Sensitivity studies were performed to determine the range of DNA concentrations that are able to produce reliable quantification results and to determine the limit of detection. The quantities of human and human male DNA obtained from the Quantifiler0003 Duo kit for the two samples were very similar to the expected quantities across a range of concentrations from 20 ng ⁄ lL to 23 pg ⁄ lL (Table 1). Furthermore, quantities as low as 11.5 pg ⁄ lL of human DNA were reproducibly detected across all replicates. At concentrations of 5.75 pg ⁄ lL and below, human DNA was not reproducibly detected across all replicates because of the stochastic variation in the amplification efficiency at low DNA input amounts. As expected, samples containing lower quantities of DNA exhibited greater variation in the quantification results because of stochastic effects. The limit of detection of human and human male DNA by the Quantifiler0003 Duo kit is similar to other real-time PCR-based assays for quantification of human and ⁄ or human DNA reported in the literature (4–8,14,15). Some of these reported assays have demonstrated a lower limit of detection than the Quantifiler0003 Duo kit. However, the ultimate goal of human and human male DNA quantification is to determine the volume of extract to be used as template for amplification using STR genotyping kits. The amount of DNA recommended for STR typing ranges from 0.5 to 2.0 ng for different kits as described in the User’s Manuals for AmpF‘STR0003 kits. In general, for samples containing DNA at concentrations of 0.1 ng ⁄ lL or less, it is necessary to add the maximum volume of DNA extract to the AmpF‘STR0003 kit amplification reaction. Therefore, quantification values of 25% difference in the quantities of human and human male DNA. Sample No. 417 34 64 129 183
Population Caucasian African-American African-American African-American African-American
Duo Human Duo Male (ng ⁄ lL) (ng ⁄ lL) 0.688 0.244 0.230 0.253 0.329
1.300 0.475 0.458 0.444 0.542
IPC CT
Male-human, % difference
29.837 29.957 29.733 29.847 29.803
88.953 94.672 99.130 75.494 64.742
IPC, internal positive control.
0003
samples. Thus, using the results generated from the Quantifiler Duo kit, it is possible to estimate which AmpF‘STR0003 kit will be likely more successful and therefore make an educated decision to choose between autosomal STR and Y-STR analysis for genotyping male DNA in a mixture sample. Population Study Genomic DNA samples from 534 individual donors of Caucasian (130 male and 60 female), African-American (116 male and 24 female), and Hispanic (129 male and 75 female) population groups were analyzed using the Quantifiler0003 Duo DNA Quantification Kit. First, the kit detected and quantified DNA in all 534
FIG. 4—Inhibitor study: CT values for RPPH1, SRY, and IPC targets for inhibited samples containing 0.5 ng ⁄ lL DNA and humic acid at final concentrations of 0, 1.0, 2.0, 3.0, 3.75, 7.5, 11.25, 15, and 30 ng ⁄ lL in the qPCR.
human DNA samples (data not shown). All male samples exhibited the SRY signal. Second, the SRY signal was not detected for any of the female samples tested. Of the 375 male samples, for 370 the quantity values for human male DNA were within €25% of the total human quantity. This range is normal and it is determined by the inherent variations in the PCR and liquid handling (pipetting). For the five remaining male samples, the kit provided male DNA quantity values that deviated from the human DNA quantity more than €25%. The results are
312
JOURNAL OF FORENSIC SCIENCES TABLE 6—Stability study: measured quantities of human and human male DNA in samples spiked with the inhibitors. SRY
Inhibitor Hematin
Humic acid
Sample Name (concentration in qPCR) 0 lM 2.5 lM 5 lM 7.5 lM 10 lM 12.5 lM 15 lM 17.5 lM 20 lM 40 lM 0 ng ⁄ lL 1 ng ⁄ lL 2 ng ⁄ lL 3 ng ⁄ lL 3.75 ng ⁄ lL 7.5 ng ⁄ lL 11.25 ng ⁄ lL 15 ng ⁄ lL 30 ng ⁄ lL
RPPH1
IPC
Quantity (ng ⁄ lL)
SD
Quantity (ng ⁄ lL)
SD
CT
SD
0.411 0.406 0.393 0.371 0.092 0.007 0.000 0.000 0.000 0.000 0.411 0.381 0.370 0.275 0.155 0.006 0.000 0.000 0.000
0.033 0.099 0.041 0.035 0.013 0.004 0.000 0.000 0.000 0.000 0.033 0.077 0.060 0.058 0.041 0.005 0.000 0.000 0.000
0.397 0.399 0.358 0.253 0.024 0.000 0.000 0.000 0.000 0.000 0.397 0.359 0.244 0.105 0.055 0.000 0.000 0.000 0.000
0.045 0.029 0.045 0.030 0.010 – – – – – 0.045 0.034 0.029 0.023 0.016 – – – –
29.647 29.833 29.677 30.050 31.753 35.347 40.000 40.000 40.000 40.000 29.647 29.783 29.813 30.263 30.807 35.207 40.000 40.000 40.000
0.112 0.078 0.123 0.122 0.377 1.048 – – – – 0.112 0.054 0.071 0.142 0.411 1.540 – – –
IPC, internal positive control; SRY, sex determining region Y; RPPH1, ribonuclease P RNA component H1.
FIG. 5—Inhibitor study: STR analysis of humic acid-inhibited samples using the Identifiler0003 kit. The sample name in the panel corresponds to the nomenclature in Table 6. The final concentration of the inhibitor in the PCR for STR varied and was determined by the volume of extract used. One nanogram of human DNA template (based on the quantification results summarized in Table 6) was used for amplification.
summarized in Table 5. Higher measured quantities of human male DNA for these samples were probably because of duplication of the SRY gene. Similar findings were previously described
and the ratio of human male DNA:human DNA (IDYZ5:AluYa5) measured for 54 males from different population groups ranged between 0.53 and 1.23 (7).
BARBISIN ET AL. • QUANTIFILER0002 DUO KIT VALIDATION
Inhibited Samples Forensic DNA extracts may contain compounds which inhibit the amplification of nucleic acids if not removed during the extraction procedures. These PCR inhibitors can interfere with the reaction and cause varying levels of reduced PCR efficiency, including complete inhibition of PCR. An IPC template is coamplified with the RPPH1 and SRY targets in the Quantifiler0003 Duo kit to monitor the presence of PCR inhibitors. Increased CT values for the IPC target indicate the extent of the presence of PCR inhibitor(s). The ability of the Quantifiler0003 Duo assay to monitor the presence of PCR inhibitors in a given sample was investigated by adding to a DNA solution hematin and humic acid, inhibitors commonly found in blood and soiled samples, respectively. The studies were conducted to determine the limit of PCR inhibitors concentration in a forensic sample that would allow relatively accurate quantification results for obtaining an interpretable STR profile. Figure 4 summarizes the CT values at different concentrations of humic acid. It is evident that the CT values were relatively stable up to 3.0 ng ⁄ lL humic acid. The PCR efficiency in the Quantifiler0003 Duo kit decreased as the concentrations of humic acid increased. Complete inhibition of the amplification occurred at 11.25 ng ⁄ lL and higher concentration of humic acid. In general, the CT values for all targets (human,
313
human male, and IPC) were affected similarly at a given concentration of inhibitor (Fig. 4). Similar results were obtained for hematin; CT values were relatively stable up to 7.5 lM hematin and increased with increasing concentrations of the inhibitor. Complete inhibition was observed at concentrations higher than 15.0 lM hematin (data not shown). The quantification results obtained from samples spiked with the inhibitors are shown in Table 6. The presence of hematin and humic acid resulted in the inhibition of PCR and adversely affected the quantification of DNA in a sample. Thus, the quantity of DNA was underestimated when the concentrations of hematin and humic acid were increased. Hematin and humic acid at concentrations higher than 12.5 lM and 7.5 ng ⁄ lL, respectively, completely inhibited the PCR so that no DNA was detectable in these samples. Using the quantification results from the RPPH1 target (Table 6), 1.0 ng of template DNA was introduced in the AmpFlSTR0003 Identifiler0003 kit PCR amplification reaction; the results for samples spiked with humic acid are presented in Fig. 5. An interpretable, complete profile was obtained for the control sample and the samples labeled as 1 and 2 ng ⁄ lL; the sample labeled as 3 ng ⁄ lL exhibited partial profiles, and all other samples did not provide any STR profiles. The results demonstrate that an upward shift in the IPC CT value with the Quantifiler0003 Duo kit can be used to predict the
FIG. 6—Inhibitor study: STR analysis of humic acid-inhibited samples before dilution of the highly inhibited samples using the MiniFilerTM kit. 0.25 ng of human DNA is used for amplification of the sample in panel (A); 0.1 ng in panels (B) and (C); 0.05 ng in panel (D); 0.03 ng in panel (E) (based on the quantification results summarized in Table 6). Ten microliter of the extract is used for amplification of the samples in panels (F–I). Few alleles in panels (F) and (G) provided off-scale data as indicated by the off-scale Peak Indicator. Sample names in the panels correspond to the nomenclature in Table 6. The final concentration of the inhibitor in the MiniFilerTM reaction varied and was determined by the volume of the extract used.
314
JOURNAL OF FORENSIC SCIENCES
FIG. 7—Inhibitor study: STR analysis of humic acid-inhibited samples after dilution of the highly inhibited samples using the MiniFilerTM kit. A total of 0.25 ng of human DNA is used for amplification of the sample in panel (A); 0.1 ng in panels (B) and (C) (based on the quantification results summarized in Table 6). Ten microliters of the diluted extract, as indicated, is used for amplification of the samples in panels (D–I). The final concentration of the inhibitor in the MiniFilerTM reaction varied and was determined by the volume of the extract used and the dilution.
nature of the STR profile that will be generated (Figs. 4 and 5). Similar results were obtained for the samples spiked with hematin: full STR profiles were obtained for the control sample and samples labeled as 2.5 lM and 5 lM, a partial profile was obtained for the sample labeled as 7.5 lM, and no profile was obtained for all the other samples (data not shown). The MiniFilerTM kit enables the recovery of STR profiles from compromised samples such as those which may be inhibited and ⁄ or TABLE 7—Stability study: measured quantities of human and human male DNA in degraded samples. SRY Sample Name 1 2 3 4 5 6 7
RPPH1
IPC
DNase I (units)
Quantity (ng ⁄ lL)
SD
Quantity (ng ⁄ lL)
SD
CT
SD
0 0.002 0.01 0.02 0.05 0.1 0.2
7.07 5.92 4.77 3.23 0.50 0.10 0.02
0.08 0.20 0.06 0.10 0.04 0.01 0.01
7.69 6.51 5.11 3.43 0.57 0.08 0.03
0.64 0.39 0.13 0.33 0.13 0.01 0.01
29.61 29.60 29.67 29.75 29.77 29.90 29.83
0.24 0.13 0.11 0.08 0.07 0.11 0.05
IPC, internal positive control; SRY, sex determining region Y; RPPH1, ribonuclease P RNA component H1.
degraded because of its design characterized by smaller amplicon lengths and improved PCR conditions (11). Figure 6 summarizes the results obtained with the MiniFilerTM kit for the samples spiked with humic acid. The control sample that did not contain any inhibitor generated a complete profiles when using 0.25 ng of template DNA for amplification. The samples labeled as 1 and 2 ng ⁄ lL provided complete profiles using 0.1 ng of DNA template. The samples labeled as 3 and 3.75 ng ⁄ lL provided complete profiles using 0.05 and 0.03 ng of DNA template, respectively. All of the other samples provided either partial or uninterpretable profiles using 10 lL of samples (low or no quantification results were obtained because of inhibition, Table 6). These results are consistent with observations of an IPC CT shift during quantification (Fig. 4). Every PCR system, e.g., Quantifiler0003 Duo, Identifiler0003, and MiniFilerTM kits, has a unique reagent formulation that provides a different response to samples containing inhibitors. When samples containing higher concentrations of humic acid were diluted (see dilution factor in Fig. 7) and 10 lL was amplified, conclusive interpretable profiles were obtained using the MiniFilerTM kit (Fig. 7). Similar results were obtained for the samples spiked with different concentrations of hematin (data not shown). Swango et al. (8) have calculated the extent of the reduction in the quantification of DNA based on the shift in the IPC CT value. This type of prediction is applicable provided only one inhibitor is present and correlation of the IPC CT values to the concentration of the
BARBISIN ET AL. • QUANTIFILER0002 DUO KIT VALIDATION
315
FIG. 8—Degraded samples study: STR analysis of artificially degraded DNA samples using the Identifiler0003 kit. The DNA was treated with increasing concentrations of DNase I: (1) 0; (2) 0.002; (3) 0.01; (4) 0.02; (5) 0.05; (6) 0.1; and (7) 0.2 DNase I Units. The rfu scale varies per panel and ranges from 900 to 4000 rfu.
FIG. 9—Degraded samples study: STR analysis of artificially degraded DNA samples using the MiniFilerTM kit. The DNA was treated with increasing concentrations of DNase I. (1) 0; (2) 0.002; (3) 0.01; (4) 0.02; (5) 0.05; (6) 0.1; and (7) 0.2 DNase I Units. A total of 0.25 ng of human DNA was amplified for samples 1–6; 0.1 ng for sample 7.
inhibitor is established, as elaborated in the study (8). In our opinion, the shift in the IPC CT provides an indication of the presence of PCR inhibitors and its correlation with the extent of underestimation of DNA quantification in forensic-type samples can become very difficult because of one or more of the following reasons: (i) it is difficult to identify the inhibitor present in the forensic sample; (ii) multiple inhibitors may be present in the sample; (iii) the inhibitor may be complexed with other compounds; and (iv) the extent of inhibition of
human, human male, and IPC targets can be affected to different extents by an inhibitor. Degraded DNA A common observation in forensic evidence samples is the fragmentation of full-length DNA molecules and the reduction of overall concentration of amplifiable DNA because of exposure to
316
JOURNAL OF FORENSIC SCIENCES
TABLE 8—Correlation study: measured and expected quantities for four male and one female DNA samples using Quantifiler0003 Duo, Human, and Y kits. Mean Quantity Human Sample Name A
B
C
D
E
0003
0003
Mean Quantity Male 0003
Expected Quantity (ng ⁄ lL)
Quantifiler Duo (ng ⁄ lL)
Quantifiler Human (ng ⁄ lL)
Mean % difference*
Quantifiler Duo (ng ⁄ lL)
Quantifiler0003 Y (ng ⁄ lL)
Mean % difference*
20.000 10.000 1.000 0.100 0.050 20.000 10.000 1.000 0.100 0.050 20.000 10.000 1.000 0.100 0.050 20.000 10.000 1.000 0.100 0.050 20.000 10.000 1.000 0.100 0.050
21.153 9.110 0.869 0.091 0.043 24.363 11.493 1.143 0.104 0.061 22.513 8.720 0.822 0.098 0.044 27.283 13.263 1.217 0.118 0.059 24.910 12.107 1.137 0.116 0.056
21.160 10.440 0.831 0.071 0.044 27.690 12.290 1.080 0.098 0.044 24.740 11.110 1.010 0.129 0.056 16.730 10.610 1.470 0.100 0.066 27.880 13.270 1.300 0.162 0.060
)0.033 )12.739 4.573 28.169 )2.273 )12.015 )6.485 5.833 6.122 38.636 )9.002 )21.512 )18.614 )24.031 )21.429 63.078 25.005 )17.211 18.000 )10.606 )10.653 )8.764 )12.538 )28.395 )6.667
20.103 8.983 0.854 0.083 0.046 23.087 11.220 1.147 0.110 0.049 23.112 9.250 0.894 0.108 0.044 26.487 13.090 1.263 0.121 0.073 Female Female Female Female Female
16.910 9.020 1.120 0.113 0.060 20.380 10.650 1.310 0.160 0.082 20.270 9.220 1.110 0.099 0.046 22.800 11.740 1.500 0.145 0.074 female female female female female
18.882 )0.410 )23.750 )26.549 )23.333 13.283 5.352 )12.443 )31.250 )40.244 14.021 0.325 )19.459 9.091 )4.348 16.171 11.499 )15.800 )16.552 )1.351 female female female female female
*Mean % difference was calculated as described in the text.
TABLE 9—Case-type sample study: measured quantities in case-type samples. Quantity (ng ⁄ lL) Sample No. 1 2 3 4 5 6 7 8
Sample Description
SRY
RPPH1
IPC CT
SRY–RPPH1 (% difference)
Saliva on cotton swab Saliva on cotton swab Blood stain on fabric Blood stain on fabric Blood stain on denim Blood stain on filter paper Blood spiked with inhibitors and stained on fabric Semen stain on fabric
2.06 11.00 0.82 2.09 1.37 1.35 1.84 1.78
2.01 11.35 0.91 2.07 0.76 1.34 1.72 1.82
29.74 29.77 29.84 29.68 32.54 29.48 29.69 29.47
2.5 )3.1 )9.7 1.0 81.0 0.7 7.0 )2.2
IPC, internal positive control; SRY, sex determining region Y; RPPH1, ribonuclease P RNA component H1.
environmental insults. A sample of high-molecular weight humangenomic DNA was used to generate a series of samples with varying levels of degradation. The quantity of DNA obtained by using the Quantifiler0003 Duo kit for the control and the samples at varying degrees of degradation is summarized in Table 7. Lower amounts of amplifiable DNA were obtained for the samples degraded with higher amounts of DNase I; the amount of human DNA decreased from about 7.69 to 3.43 ng ⁄ lL when 0.02 units of DNase I were used, and to 0.03 ng ⁄ lL when 0.2 units of DNase I were used according to the results provided by the human target RPPH1. The values obtained from the SRY target assay were very similar. The correlation between the DNA quantity in the degraded samples and STR profiles generated for Identifiler0003 and MiniFilerTM was investigated. Using the RPPH1 target quantification results, 1.0 ng of DNA or 10 lL of the extract (for samples that exhibited
The quantity of DNA was determined by Quantifiler Duo, Quantifiler Human, and Quantifiler Y Human Male DNA Quantification Kits (Applied Biosystems). Real-Time PCR Amplification Real-time PCR amplification reactions contained 10.5 lL of Primer-Probe Mix, 12.5 lL of Master Mix, and 2.0 lL of DNA sample.
Abstract
Analysis of the length polymorphisms of short tandem repeats (STR) loci in the human genome has become a standard approach for comparative genotyping in many areas including disease research and diagnostics, parentage assessment, investigations of human diversity, and forensic science. The purpose of this study is to optimize the DNA concentration in ng/10μL for amplification of DNA markers. AmpFlSTR Identifiler Kit is used to amplify STR markers and capillary electrophoresis is used to analyze DNA profile of human the genome. Two sets of samples with following DNA concentration: 100 pg – 6 ng/25 μL were used for this study. There was no DNA profile detected in samples with concentrations 100 pg - 300 pg/25 μL (pictogram), while in some cases partial DNA profile was yielded. On the other hand samples with 0.4 ng – 4 ng/25 μL, yielded a full DNA profile. We were not able to obtain any profile using concentrations over 4 ng/25 μL. Improvements in detection limits/sensitivity at upper and lower DNA concentrations are of potential benefits to amplify STR of Human Genomic in order to obtain a full DNA profile. The optimal DNA concentrations which produced reliable and balanced peaks, no off scale peaks and full DNA profile for all loci were at range 0.4 ng – 3 ng/25 μL.
INTRODUCTION
Eukaryotic chromosomal DNA, polymorphic short tandem repeat (STR) loci are key tools for: rapid gene discovery, disease locus mapping and carrier diagnosis of disease states, linkage analyses, agricultural genetics, parentage assessment, and population diversity studies. The ability to detect genetic differences between individuals increases when DNA typing information at multiple polymorphic STR loci is combined [1, ]. Clinical diagnostic laboratories usually perform analyses of biological samples that have been collected and stored in ideal conditions, making sample quality and quantity rarely an issue. Those ideal conditions are usually not met in the DNA typing analysis of forensic biological evidence, as there is no control over the amount of biological material left at a crime scene or its level of degradation or contamination due to exposure to various environmental insults [3, ]. Forensic biological samples may present challenges for DNA typing analysis, even with the utmost care throughout crime scene evidence recovery and storage. Due to potential exposure to a virtually unlimited number of uncontrolled variables, forensic casework specimens may result in particularly challenging polymerase chain reaction (PCR) templates. Optimized concentration of DNA and reagents is very important on PCR amplification of different combinations of 15 polymorphic tetranucleotide STR loci in a single reaction tube. This study has done to check the upper and lower limits of DNA concentration used in amplification step to obtain full DNA profiles [5].
MATERIAL AND METHODS
Samples
Our study was performed using human blood as biological material. Before analysis the blood sample has dried on filter paper. The size of the samples used for extraction were 3×3 mm2 and Chelex-100 resin is used as extraction method [6].
Procedures
For quantification purposes Absolute Quantitation method is used, real-time quantitative PCR assay with a fluorogenic TaqMan® probe targeting the human telomerase reverse transcriptase gene (hTERT) and supported by Quantifiler® Human DNA Quantification Kit on Instrument ABI PRISM® 7000 Sequence Detection System (Applied Bioystems-AB, P/N 4330087 with SDS Software v1.0) [7, 8]. For amplification purposes we introduced AmpFlSTR® Identifiler® PCR Amplification Kit (AB P/N 4322288) [5]. The following concentrations (ng/10 μL) of template DNA were amplified in a total reaction volume of 25 μl: 0.1, 0.2, 0.3 up to 6 ng. Samples were amplified in 0.2-ml microAmp reaction tubes with caps (AB P/N N801-0540) using the following conditions (recommended by the manufacturer, Applied Biosystems): 95°C for 11 min, 28 cycles of 94°C for 1 min, 59°C for 1 min and 72°C for 1 min, 60°C extension for 60 min and 2-8°C until samples were analyzed further. The amplification was carried out on GeneAmp PCR System 9700 (AB P/N N805-0001) [5], [1]. The amplified DNA markers were: D8S1179, D21S11, D7S820, CSF1P0, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, Amelogenin, D5S818 and FGA [5], [1]. Finally detection of amplified samples was done using 1.5 μl of amplified DNA product that was combined with aliquot 25 μL of the Hi-Di™ Formamide (AB P/N 4311320) mixed with GeneScan™ 500 LIZ™ Size Standard (AB P/N 4322682). Capillary electrophoresis was carried out using an automated DNA sequencer ABI PRISM® 310 Genetic Analyzer, AB P/N 310-00-200/240-W. The raw data were analyzed using GeneMapper® ID Software v3.2. The minimum peak height threshold was set at 50 RFU [9, 10].
RESULTS
Samples with DNA concentration at 0.1 and 0.2 ng/25 μL yielded no DNA profile (Figure 1). No DNA profile was obtained from samples with 0.3 ng/25μL, however in this case some loci were included (Figure 2). Baseline had noisy and allelic imbalance as well. Samples with DNA concentration from 0.4 - 3 ng/25μL yielded full DNA profiles (Figure 3). There were no any allelic imbalance, stutter or any other artifacts except for the sample concentration at 3 ng/25μL. Samples with concentrations 2.7 and 3 ng/25μL exhibit decrease of Relative Fluorescence Units (RFU) starting from shorter loci toward longer ones for all panels. The loci with lower RFU were D7S8210, CSF1PO, D2S1338, D18S51 and FGA [11].
Presents electropherogram of sample with No DNA Profile analyzed with AmpFlSTR Identifiler Kit. There is no any allele obtained in total of 16 human genome loci analyzed.
Presents electropherogram of samples with Partial DNA Profile analyzed with AmpFlSTR Identifiler Kit. There are three alleles obtained in total of 16 human genome loci analyzed.
Presents electropherogram of samples with Full DNA Profile analyzed with AmpFlSTR Identifier Kit. There are all alleles obtained in total of 16 human genome loci analyzed.
DNA concentration between 3 - 4 ng/25 μL yielded full DNA profiles but there were allelic imbalance peaks and significant difference on RFU in heterozygote peak height ratios between the shorter and longer loci (Figure 4).
Presents electropherogram of samples with Full DNA Profile analyzed with AmpFlSTR Identifier Kit. There are all alleles obtained in total of 16 human genome loci analyzed. At some alleles there are some imbalance peaks.
Quantifiler Human Dna Quantification Kits User’s Manual
For shorter loci RFU was over 1500 to 3500, whereas for longer ones were under 500. Despite these results, DNA profile obtained from above samples can be used officially as DNA Profile at System of Justice. No DNA profile was yielded in samples over 4 ng/25μL (Figure 1). Some loci were obtained, but just few of them (below 5) so they did not represent any relevance in DNA profile. To be valid the DNA profile must have at least 10 loci obtained. Those profiles with number of loci below 10 are unavailable to be used for any purpose [1], [9]. The referent samples yielded a full DNA profile. The best DNA profile was obtained from samples with DNA concentration at 0.5 – 1.8 ng/25 μL. Reliable DNA profiles were obtained from samples with 0.4, 0.5 and 2 – 4 ng/25 μL as well (Table 1).
TABLE 1
Represents DNA profile yielded on different DNA concentrations

DISCUSSION
A variety of commercial Kits for DNA amplification are now available and they routinely are used in forensic and diagnostic laboratories worldwide. Although generally reliable and convenient, many of these Kits offer only a relatively limited dynamic range for upper and lower limits, so that in many cases samples from the crime scene, reference samples and clinical samples frequently have to be diluted or concentrated and retested in order to avoid exceedi ng the upper or lower limits. At the same time analyzes is performed by using real-time PCR which is time and money consuming. The advantage of the current study is the fact that we found that upper and lower limits beyond of what is actually suggested by companies. This method increases the opportunity to amplify DNA directly from extraction step into amplification one without any trouble toward result interpretation. DNA yielded profile from an amount up to 4 ng/25 μL (which is a double amount suggested by protocols), crucial due to the fact that is easy to determine that approximate amount in our sample size during the extraction step. This decreases a workflow of our analysis which in this case will consist of extraction, amplification and detection without quantification step. This workflow would not have any interference in terms of detecting sensitivity of DNA. In addition, this method can serve as a great tool to help laboratories in obtaining reliable DNA profiles by saving time and money. Leclair et al. have studied lower and upper limits for DNA concentrations and total volume used for DNA amplification. In this study the best results were yielded using 0.5 ng of DNA in a 10-μL PCR reaction volume which appeared to produce an identical profile to 2 ng of the same DNA in a 40-μL reaction volume []. Company Applied Biosystems which has performed a validation of the AmpFlSTR Identifiler Kit, suggested using an amount of DNA concentration in the range of 0.5 - 2 ng/25 μL [5].
CONCLUSIONS
AmpFlSTR Identifiler, the PCR Amplification Kit used to amplify STR markers of analyzed samples exhibited to be very stable. Primers of this Kit were sensitive and very specific for DNA markers as mentioned above. Even if AmpFlSTR Identifiler PCR Amplification Kit – User manual suggests using DNA concentration 0.5 – 1.2 ng/25 μL (lower and upper limits) [5], and National Institute of Standards and Technology (NIST) suggests 0.5 – 2 ng/25 μ L as the best range of STR Kits [1]. We have obtained full DNA profile with a lower concentration of 0.4 ng/25 μL up to 4 ng/25 μL for the upper one. Other reagents of this Kit: AmpFlSTR® PCR Reaction Mix, AmpliTaq Gold® DNA Polymerase and AmpFlSTR® Identifiler™ Allelic Ladder used on 15μL as a master mix shown to be enough for amplification of samples with concentration 4 ng/25μL without any problem for inhibition by the template. Performing the step of DNA quantification on RT-PCR is time consuming and cost effective. We found that the best and reliable results are yielded beyond the limits suggested by different companies. We obtained reliable DNA profile from concentration up to 4 ng per sample analyzed. Using those results, the laboratories can make validation of methods in different applications such as: molecular biology, genetics, biomedicine etc to analyze samples directly from extraction to amplification and by skipping quantification step in RT-PCR. By applying such approach it could be possible to save time and money.
ACKNOWLEDGMENT
We would like to thank you Nancy Whitney Peterson (Forensic Biology/DNA Consultants, LLC, USA) for constructive criticism and suggestions in the preparation of the manuscript.
DECLARATION OF INTEREST
The authors have no conflict of interest to declare.